Bidisperse simulations¶
In this notebook we will show how to do simulations with different types of particles. The same principle should be applied for simulations with several types of traps.
[1]:
import os
import sys
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import matplotlib as mpl
sys.path.insert(0, '../../')
import magcolloids as mgc
from IPython.display import HTML
idx = pd.IndexSlice
[2]:
%reload_ext autoreload
%autoreload 2
[3]:
ureg = mgc.ureg
Several types of particles:¶
A particles object creates one type of particles, with a fixed set of parameters. In the simulation, this particle is copied many times to the positions given by positions.
To create a simulation with several types of particles, it is necessary to create different instances of particles. The sim and world objects accept an array of particles as arguments.
[4]:
region = np.array([20,20,4.1])
[5]:
particles = mgc.particles(
np.array([[-5,0,0],[0,5,0],[5,0,0]])*ureg.um,
radius = 2*ureg.um,
susceptibility = 1,
diffusion=0.07*ureg.um**2/ureg.s,
density = 0*ureg.kg/ureg.m**3,
temperature=300*ureg.K)
particles2 = mgc.particles(
np.array([[-5,-3,0],[0,-3,0],[5,-3,0],[5,5,0]])*ureg.um,
radius = 1*ureg.um,
susceptibility = 1,
diffusion=0.07*ureg.um**2/ureg.s,
density = 0*ureg.kg/ureg.m**3,
temperature=300*ureg.K)
[6]:
field = mgc.field(magnitude = 3*ureg.mT, frequency = 100*ureg.Hz, angle = 90*ureg.degrees)
world = mgc.world([particles,particles2], temperature = 300*ureg.K,
region=region*ureg.um, boundaries = ['p','p','f'], walls = [False,False,True],
dipole_cutoff = 20*ureg.um)
sim = mgc.sim(dir_name = "bidisperse/", file_name = "test_particles",
timestep = 1e-4*ureg.s, framerate = 30*ureg.Hz, total_time = 30*ureg.s,
particles = [particles,particles2], world = world, field = field,
output = ["x","y","z","mux","muy","muz"])
[7]:
sim.generate_scripts()
sim.run()
sim.load()
[8]:
trj = sim.lazy_read[:]
[9]:
plt.figure(figsize=(3,3),dpi=150)
ax = mgc.draw_trj(trj,sim,cmap="plasma")
for i, trj_i in trj.groupby("id"):
if all(trj_i.type==1):
ax.plot(trj_i.x,trj_i.y, color="red",linewidth = 0.5)
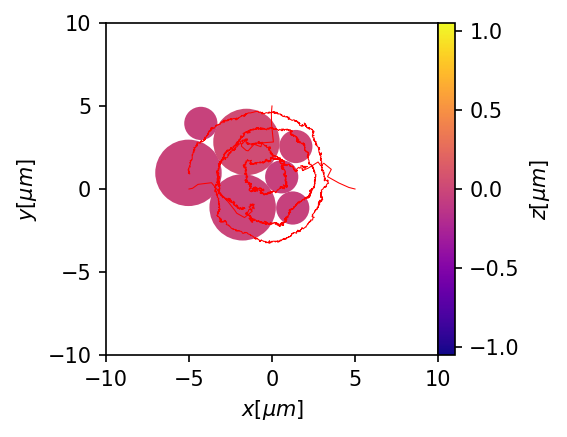
Bidisperse traps¶
Just as particles object, the bistable_trap object creates a type of trap. Here as well, we must create different types of traps to give them different parameters.
However, we must also specify the particles that will be affected by each trap. This is done by the particles argument. If we want a specific bistable_trap object to act only on some of the copies of the object particles, we can give it a subset specification. The subset can be a slice, or an array of indices which points to the particle in position \(i\) inside the particles object.
[11]:
region = np.array([20,20,4.1])
[12]:
particles = mgc.particles(
np.array([[-5,0,0],[0,5,0],[5,0,0]])*ureg.um,
radius = 2*ureg.um,
susceptibility = 1,
diffusion=0.07*ureg.um**2/ureg.s,
density = 0*ureg.kg/ureg.m**3,
temperature=300*ureg.K)
traps = mgc.bistable_trap(
np.array([[-5,0,0],[0,5,0]])*ureg.um,
np.array([[1,0,0],[0,1,0]]),
particles, subsets = slice(0,2),
distance = 2*ureg.um,
height = 0 * ureg.pN*ureg.nm,
stiffness = 3e-4 * ureg.pN/ureg.nm)
traps2 = mgc.bistable_trap(
np.array([[5,0,0]])*ureg.um,
np.array([[1,0,0]]),
particles, subsets = [2],
distance = 0*ureg.um,
height = 0 * ureg.pN*ureg.nm,
stiffness = 3e-4 * ureg.pN/ureg.nm)
[13]:
field = mgc.field(magnitude = 0*ureg.mT, frequency = 100*ureg.Hz, angle = 90*ureg.degrees)
world = mgc.world(particles, [traps,traps2], temperature = 300*ureg.K,
region=region*ureg.um, boundaries = ['p','p','f'], walls = [False,False,True],
dipole_cutoff = 20*ureg.um)
sim = mgc.sim(dir_name = "bidisperse/", file_name = "test_traps",
timestep = 1e-4*ureg.s, framerate = 30*ureg.Hz, total_time = 60*ureg.s,
particles = particles,traps = [traps,traps2], world = world, field = field,
output = ["x","y","z","mux","muy","muz"])
[14]:
sim.generate_scripts()
sim.run()
sim.load()
trj = sim.lazy_read[:]
[15]:
plt.figure(figsize=(3,3),dpi=150)
ax = mgc.draw_trj(trj[trj.type==1],sim,cmap="plasma")
for i, trj_i in trj.groupby("id"):
if all(trj_i.type==1):
ax.plot(trj_i.x,trj_i.y, color="red")
else:
ax.plot(trj_i.x,trj_i.y,'.',color="k", linewidth = .5)
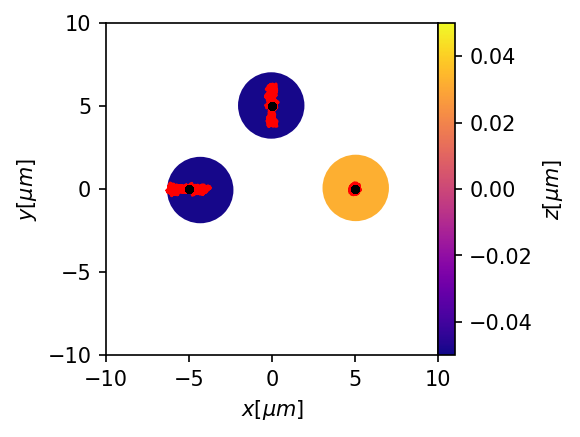
What happens if some of the traps are pair traps?¶
Pair traps are not assigned a bond to a specific trap, but instead they act on whatever particles are inside their cutoff (pair traps must have a cutoff).
[16]:
region = np.array([20,20,4.1])
[17]:
particles = mgc.particles(
np.array([[-5,0,0],[0,5,0],[5,0,0]])*ureg.um,
radius = 2*ureg.um,
susceptibility = 1,
diffusion=0.07*ureg.um**2/ureg.s,
density = 0*ureg.kg/ureg.m**3,
temperature=300*ureg.K)
traps = mgc.bistable_trap(
np.array([[-5,0,0],[0,5,0]])*ureg.um,
np.array([[1,0,0],[0,1,0]]),
particles, subsets = slice(0,2),
distance = 2*ureg.um,
height = 0 * ureg.pN*ureg.nm,
stiffness = 3e-4 * ureg.pN/ureg.nm)
traps2 = mgc.bistable_trap(
np.array([[5,0,0]])*ureg.um,
np.array([[1,0,0]]),
particles, subsets = [2],
distance = 0*ureg.um,
height = 0 * ureg.pN*ureg.nm,
stiffness = 1e-4 * ureg.pN/ureg.nm,
cutoff = 1*ureg.um)
[18]:
field = mgc.field(magnitude = 0*ureg.mT, frequency = 100*ureg.Hz, angle = 90*ureg.degrees)
world = mgc.world(particles, [traps,traps2], temperature = 300*ureg.K,
region=region*ureg.um, boundaries = ['p','p','f'], walls = [False,False,True],
dipole_cutoff = 20*ureg.um)
[19]:
sim = mgc.sim(dir_name = "bidisperse/", file_name = "test_traps_pair",
timestep = 1e-4*ureg.s, framerate = 30*ureg.Hz, total_time = 60*ureg.s,
particles = particles,traps = [traps,traps2], world = world, field = field,
output = ["x","y","z","mux","muy","muz"])
[20]:
sim.generate_scripts()
sim.run()
sim.load()
trj = sim.lazy_read[:]
[21]:
plt.figure(figsize=(3,3),dpi=150)
ax = mgc.draw_trj(trj[trj.type==1],sim,cmap="plasma")
for i, trj_i in trj.groupby("id"):
if all(trj_i.type==1):
ax.plot(trj_i.x,trj_i.y, color="red")
else:
ax.plot(trj_i.x,trj_i.y,'.',color="k", linewidth = .5)
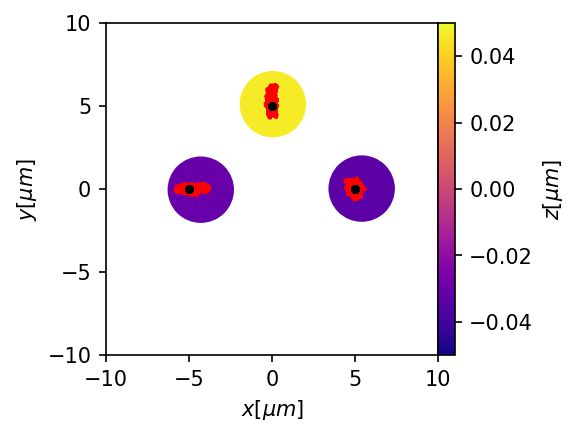
This also works well. In principle, a single particle should be able to be traped by several traps if they have a cutoff. Let’s see if that works.
[22]:
region = np.array([20,20,4.1])
[23]:
particles = mgc.particles(
np.array([[-2.5,-5,0],[0,5,0],[2.5,-5,0]])*ureg.um,
radius = 2*ureg.um,
susceptibility = 0,
diffusion=0.07*ureg.um**2/ureg.s,
density = 0*ureg.kg/ureg.m**3,
temperature=300*ureg.K)
traps = mgc.bistable_trap(
np.array([[-1,-5,0],[0,5,0]])*ureg.um,
np.array([[1,0,0],[0,1,0]]),
particles, subsets = slice(0,2),
distance = 2*ureg.um,
height = 0 * ureg.pN*ureg.nm,
stiffness = 3e-4 * ureg.pN/ureg.nm)
traps2 = mgc.bistable_trap(
np.array([[1,-5,0]])*ureg.um,
np.array([[1,0,0]]),
particles, subsets = [2],
distance = 0*ureg.um,
height = 0 * ureg.pN*ureg.nm,
stiffness = 1e-6 * ureg.pN/ureg.nm,
cutoff = 10*ureg.um)
[24]:
field = mgc.field(magnitude = 0*ureg.mT, frequency = 100*ureg.Hz, angle = 90*ureg.degrees)
world = mgc.world(particles, [traps,traps2], temperature = 300*ureg.K,
region=region*ureg.um, boundaries = ['p','p','f'], walls = [False,False,True],
dipole_cutoff = 20*ureg.um)
[25]:
sim = mgc.sim(dir_name = "bidisperse/", file_name = "test_traps_pair",
timestep = 1e-4*ureg.s, framerate = 30*ureg.Hz, total_time = 60*ureg.s,
particles = particles,traps = [traps,traps2], world = world, field = field,
output = ["x","y","z","mux","muy","muz"])
[26]:
sim.generate_scripts()
sim.run()
sim.load()
trj = sim.lazy_read[:]
[27]:
plt.figure(figsize=(3,3),dpi=150)
ax = mgc.draw_trj(trj[trj.type==1],sim,cmap="plasma")
for i, trj_i in trj.groupby("id"):
if all(trj_i.type==1):
ax.plot(trj_i.x,trj_i.y, color="red")
else:
ax.plot(trj_i.x,trj_i.y,'.',color="k", linewidth = .5)
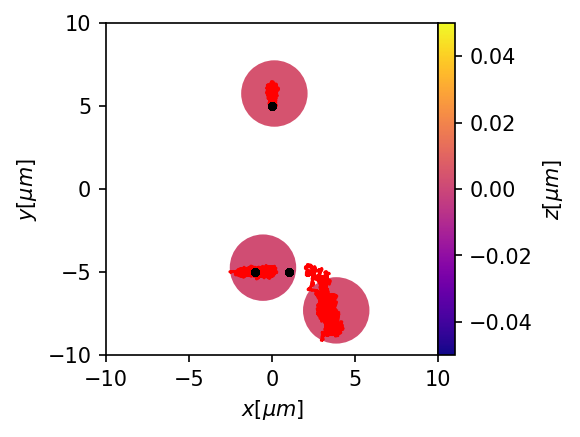
This works, but keep in mind that all particles see the pair trap. Currently, I don’t know how to make some particles see the pair trap and some not. Maybe this can be implemented in the future by defining several particle types.
Bidisperse traps and bidisperse particles¶
If the list of particle objects particles, has \(N_p\) particle types, and there are \(N_t\) traps of type \(t\), subsets should be a list of \(N_t\) pairs. The pair p = subsets[i] indicates trap \(i\) is to be bonded with particle \(p[1]\) in the list particles[p[0]].
[28]:
large_particles = mgc.particles(
np.array([[0,2.5,0]])*ureg.um,
radius = 2*ureg.um,
susceptibility = 0,
diffusion=0.07*ureg.um**2/ureg.s,
density = 0*ureg.kg/ureg.m**3,
temperature=300*ureg.K)
small_particles = mgc.particles(
np.array([[-2.5,0,0],[0,0,0]])*ureg.um,
radius = 1*ureg.um,
susceptibility = 0,
diffusion=0.07*ureg.um**2/ureg.s,
density = 0*ureg.kg/ureg.m**3,
temperature=300*ureg.K)
## The trap at [-2.5,0,0] should be paired with the first small particle.
## The trap at [2.5,0,0] should be paired with the first large particle.
long_traps = mgc.bistable_trap(
np.array([[-2.5,0,0],[0,2.5,0]])*ureg.um,
np.array([[1,0,0],[0,1,0]]),
[large_particles,small_particles], subsets = [[1,0],[0,0]],
distance = 2*ureg.um,
height = 0 * ureg.pN*ureg.nm,
stiffness = 3e-4 * ureg.pN/ureg.nm)
## The trap at [0,0,0] should be paired with the second small particle.
short_trap = mgc.bistable_trap(
np.array([[0,0,0]])*ureg.um,
np.array([[1,0,0]]),
[large_particles,small_particles], subsets = [[1,1]],
distance = 0*ureg.um,
height = 0 * ureg.pN*ureg.nm,
stiffness = 3e-4 * ureg.pN/ureg.nm)
[29]:
field = mgc.field(magnitude = 0*ureg.mT, frequency = 100*ureg.Hz, angle = 90*ureg.degrees)
world = mgc.world([large_particles,small_particles],
[long_traps, short_trap], temperature = 300*ureg.K,
region=region*ureg.um, boundaries = ['p','p','f'],
walls = [False,False,True],
dipole_cutoff = 20*ureg.um)
[30]:
sim = mgc.sim(dir_name = "bidisperse/", file_name = "test_traps_bibi",
timestep = 1e-4*ureg.s, framerate = 30*ureg.Hz, total_time = 60*ureg.s,
particles = [large_particles,small_particles],
traps = [long_traps, short_trap], world = world, field = field,
output = ["x","y","z","mux","muy","muz"])
[33]:
sim.generate_scripts()
sim.run()
sim.load()
trj = sim.lazy_read[:]
[34]:
plt.figure(figsize=(3,3),dpi=150)
ax = mgc.draw_trj(trj.query("type<3"),sim,cmap="plasma")
for i, trj_i in trj.groupby("id"):
if all(trj_i.type<3):
ax.plot(trj_i.x,trj_i.y)
else:
ax.plot(trj_i.x,trj_i.y,'.',color="k", linewidth = .5)
This last part probably breaks the behaviour above